Data, Directly

F&I researcher Julie Schram extracts lipids in her laboratory, located at Lena Point Fisheries Facility in Juneau.
By Joey Hogenson
Alaska NSF EPSCoR
Fire & Ice researchers Julie Schram and Jessica Glass are doing things the easy way.
At least, that’s the idea. Schram, a Co-PI (and a faculty hire) of the Alaska NSF EPSCoR Fire & Ice project and an Assistant Professor of Animal Physiology at the University of Alaska Southeast, and Glass, a fellow Fire & Ice faculty hire and an Assistant Professor in the UAF College of Fisheries and Ocean Sciences, are using biochemical lab techniques that enable them to collect data about marine life and their interactions without having to directly observe them in real time.

Julie Schram freeze-dries biomass samples in her lab.
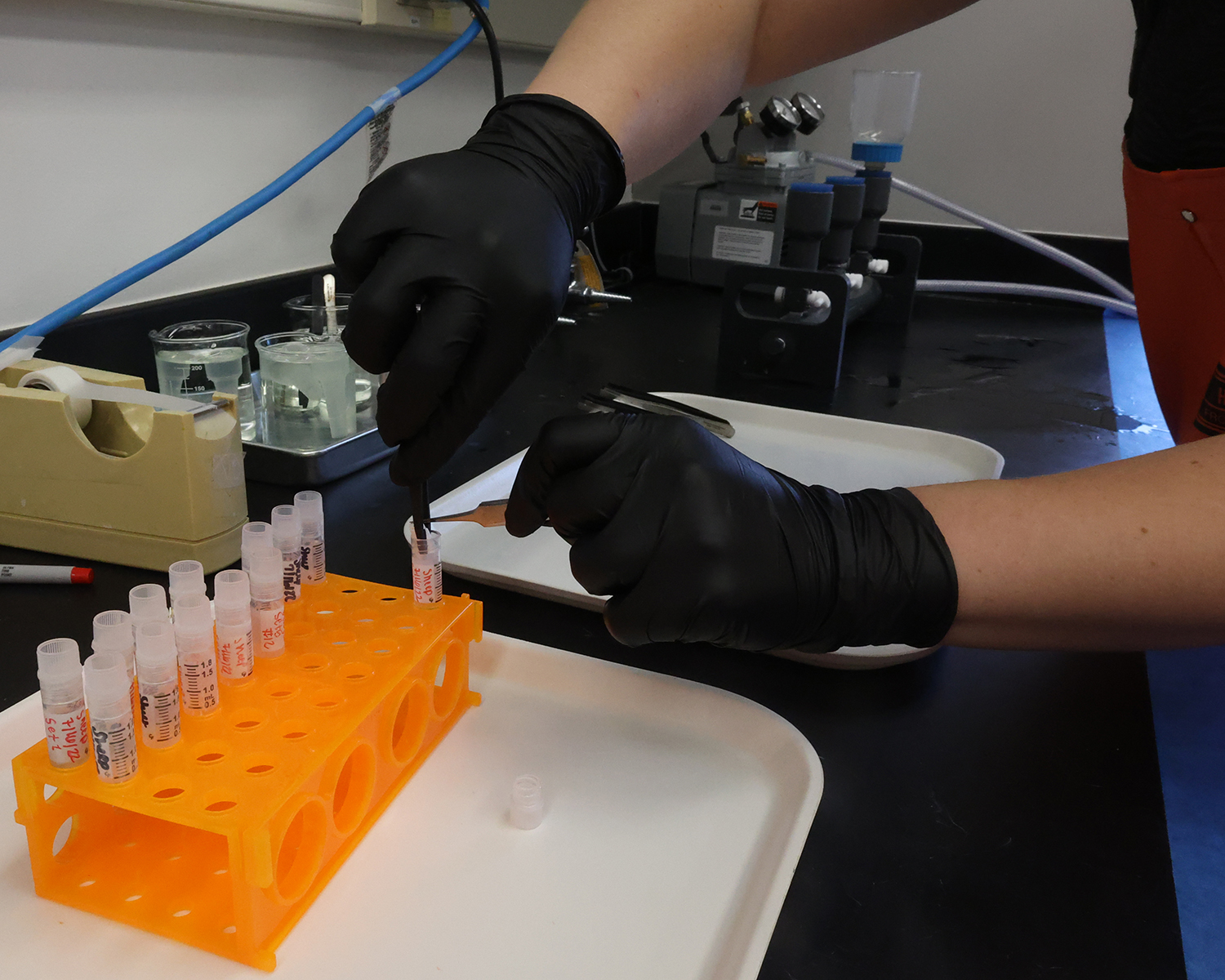
Schram’s students and colleagues are collecting marine invertebrates and seaweeds each month at selected Kachemak Bay and Juneau-area watersheds. They then freeze-dry the samples and crush them into a fine powder before extracting lipids from the organisms, which will be analyzed to gain a better understanding of the respective organisms’ diets and their overall condition.

(l to r) UAF Master’s student Maris Goodwin and UAS undergraduate Gracelyn Ham gather water samples for eDNA research near Juneau.
This information will be linked to environmental sensor data collected at the same sites and compared with stable isotope data collected by F&I researcher Katrin Iken and her students, helping us to increase our understanding of how environment influences organisms’ conditions.

Goodwin filters water samples to retrieve sediment for eDNA testing.
Additionally, Schram is working with UAF postdoctoral researcher Scotty Gabara and UAS undergraduate Gracelyn Ham on projects that combines field and laboratory experiments to better understand how glacial melt affects seaweed nutritional quality and how that may influence different aspects of intertidal marine invertebrates. Gabara conducted a lab experiment during which he measured the feeding rates of periwinkles and limpets collected from sites with different glacier extents over five months. His work will help us to better understand how the amount of glaciation of a watershed impacts the nutritional quality of the seaweeds consumed by these herbivorous invertebrates. Ham’s experiment focuses on how glacial melt may affect blue mussels, in terms of both their nutritional condition and aspects of their performance like adhesion to rock surfaces and shell strength.

Goodwin folds the eDNA filter for transport in vials.
In conjunction with Schram’s research, Glass’ research team is collecting water samples from the same coastal sites and analyzing their environmental DNA (eDNA), which is DNA found free-floating in the water. This powerful technique enables researchers to measure biodiversity across the Tree of Life, from fishes to invertebrates to seabirds and mammals, without having to collect each and every specimen. The team collects and filters ocean water, then uses Polymerase Chain Reaction (PCR) metabarcoding techniques to amplify DNA found in the sample. Once tests are completed they show what species’ DNA was present in the water and in what amounts, shedding light on biodiversity at the sampling site.
Ham places a filter in a vial for eDNA analysis.
These innovative techniques have the potential to significantly expedite research into marine biodiversity and food webs, increasing our knowledge about northern marine environments undergoing climate-related changes.

